Check out p-ter's post at Gene Expression about a meeting report of "Human Genome Variation 2006" in Hong Kong which it looks like is a very exclusive meeting limited to 200 people. There are some interesting quotes that p-ter highlights in his post about gradients of allele frequencies in the US and differences in genetic patterns (LD and heterozygosity) between HapMap Caucasians and Ashkenazi subjects. Greg Cochran must be loving it.
I would also add that the meeting also highlighted the importance of non-coding regions as being responsible for pathologies, and for differing levels of gene expression.
Wednesday, January 31, 2007
Monday, January 29, 2007
Hominin diversity

I found this article from Natural History Magazine via the Anthropology in the News website from Texas A&M Univeristy (my undergrad alma mater). It deals with reconstructing faces of the human past. It struck me how diffferent and diverse hominins were and how this much diversity happened only within the 3-4 million years.
The paper discusses how they reconstruct the faces from the existing bone structure of the skull. Seeing the diversity of faces makes me
 think of whether in our EEA (environment of evolutionary adaptedness) we often encountered members of different hominin species. The article mentions that there is some evidence of this. A common argument used be evolutionary psychologists against studying ethnicity/race as an evolutionary based mechanism is that the presence of different forms of humans would have been a novel experience for us now in the modern world. Before we never would have experienced phenotypically different humans. I have seen it suggested in this article and elsewhere that this is a possibility.
think of whether in our EEA (environment of evolutionary adaptedness) we often encountered members of different hominin species. The article mentions that there is some evidence of this. A common argument used be evolutionary psychologists against studying ethnicity/race as an evolutionary based mechanism is that the presence of different forms of humans would have been a novel experience for us now in the modern world. Before we never would have experienced phenotypically different humans. I have seen it suggested in this article and elsewhere that this is a possibility.Check out a post from a few months ago about a paper that disputes this possibility.


Looking at these pictures also highlights to me the fact that the path to modern humans was a well worn one- with respect to different diets, physiology, social systems ... all as recently as 30,000 years ago, with Neanderthals.
Argument against "Aquatic Ape"
 Basically, according to them, there is evidence that we could have obtained sufficient amounts of good brain fats from a terrestrial ecosystem. Not sure what this evidence consists of -- I don't have access to the full-text of this paper.
Basically, according to them, there is evidence that we could have obtained sufficient amounts of good brain fats from a terrestrial ecosystem. Not sure what this evidence consists of -- I don't have access to the full-text of this paper.Docosahexaenoic acid, the aquatic diet, and hominin encephalization: Difficulties in establishing evolutionary links
Bryce A. Carlson, John D. Kingston
American Journal of Human Biology Volume 19, Issue 1 , Pages 132 - 141
Abstract: Distinctive characteristics of modern humans, including language, tool manufacture and use, culture, and behavioral plasticity, are linked to changes in the organization and size of the brain during hominin evolution. As brain tissue is metabolically and nutritionally costly to develop and maintain, early hominin encephalization has been linked to a release of energetic and nutritional constraints. One such nutrient-based approach has focused on the n-3 long-chained polyunsaturated fatty acid docosahexaenoic acid (DHA), which is a primary constituent of membrane phospholipids within the synaptic networks of the brain essential for optimal cognitive functioning. As biosynthesis of DHA from n-3 dietary precursors (alpha-linolenic acid, LNA) is relatively inefficient, it has been suggested that preformed DHA must have been an integral dietary constituent during evolution of the genus Homo to facilitate the growth and development of an encephalizing brain. Furthermore, preformed DHA has only been identified to an appreciable extent within aquatic resources (marine and freshwater), leading to speculation that hominin encephalization is linked specifically to access and consumption of aquatic resources. The key premise of this perspective is that biosynthesis of DHA from LNA is not only inefficient but also insufficient for the growth and maturation demands of an encephalized brain. However, this assumption is not well-supported, and much evidence instead suggests that consumption of LNA, available in a wider variety of sources within a number of terrestrial ecosystems, is sufficient for normal brain development and maintenance in modern humans and presumably our ancestors.
Fetal Origin (or DOHaD) vs. Thrifty Gene
This is the typical alternative explanation to the thrifty genotype hypothesis for our metabolic syndrome. I particularly like this passage, from the abstract:
"In mammals, an adverse intrauterine environment results in an integrated suite of responses, suggesting the involvement of a few key regulatory genes, that resets the developmental trajectory in expectation of poor postnatal conditions. Mismatch between the anticipated and the actual mature environment exposes the organism to risk of adverse consequences - the greater the mismatch, the greater the risk. For humans, prediction is inaccurate for many individuals because of changes in the postnatal environment toward energy-dense nutrition and low energy expenditure, contributing to the epidemic of chronic noncommunicable disease."
also see this previous post on a paper challenging the Thrifty Gene Hypothesis
Early life events and their consequences for later disease: A life history and evolutionary perspective
Peter D. Gluckman, Mark A. Hanson, Alan S. Beedle
American Journal of Human Biology Vol. 19, Issue 1, Pages 1-19
Abstract: Biomedical science has little considered the relevance of life history theory and evolutionary and ecological developmental biology to clinical medicine. However, the observations that early life influences can alter later disease risk - the developmental origins of health and disease (DOHaD) paradigm - have led to a recognition that these perspectives can inform our understanding of human biology. We propose that the DOHaD phenomenon can be considered as a subset of the broader processes of developmental plasticity by which organisms adapt to their environment during their life course. Such adaptive processes allow genotypic variation to be preserved through transient environmental changes. Cues for plasticity operate particularly during early development; they may affect a single organ or system, but generally they induce integrated adjustments in the mature phenotype, a process underpinned by epigenetic mechanisms and influenced by prediction of the mature environment. In mammals, an adverse intrauterine environment results in an integrated suite of responses, suggesting the involvement of a few key regulatory genes, that resets the developmental trajectory in expectation of poor postnatal conditions. Mismatch between the anticipated and the actual mature environment exposes the organism to risk of adverse consequences - the greater the mismatch, the greater the risk. For humans, prediction is inaccurate for many individuals because of changes in the postnatal environment toward energy-dense nutrition and low energy expenditure, contributing to the epidemic of chronic noncommunicable disease. This view of human disease from the perspectives of life history biology and evolutionary theory offers new approaches to prevention, diagnosis and intervention.
"In mammals, an adverse intrauterine environment results in an integrated suite of responses, suggesting the involvement of a few key regulatory genes, that resets the developmental trajectory in expectation of poor postnatal conditions. Mismatch between the anticipated and the actual mature environment exposes the organism to risk of adverse consequences - the greater the mismatch, the greater the risk. For humans, prediction is inaccurate for many individuals because of changes in the postnatal environment toward energy-dense nutrition and low energy expenditure, contributing to the epidemic of chronic noncommunicable disease."
also see this previous post on a paper challenging the Thrifty Gene Hypothesis
Early life events and their consequences for later disease: A life history and evolutionary perspective
Peter D. Gluckman, Mark A. Hanson, Alan S. Beedle
American Journal of Human Biology Vol. 19, Issue 1, Pages 1-19
Monday Map
I've decided to start a new weekly feature called Monday Maps (of course, I had to use the trendy alliteration). Every Monday I will display some kind of map. From Sunwize.com, this world insolation map "shows the amount of solar energy in hours, received each day on an optimally tilted surface during the worst month of the year."




Friday, January 26, 2007
Synonymous SNPs not so "silent"
A "Silent" Polymorphism in the MDR1 Gene Changes Substrate Specificity
Science 26 January 2007 Vol. 315. no. 5811, pp. 525 - 528
Chava Kimchi-Sarfaty, Jung Mi Oh, In-Wha Kim, Zuben E. Sauna, Anna Maria Calcagno, Suresh V. Ambudkar, Michael M. Gottesman
Abstract: Synonymous single-nucleotide polymorphisms (SNPs) do not produce altered coding sequences, and therefore they are not expected to change the function of the protein in which they occur. We report that a synonymous SNP in the Multidrug Resistance 1 (MDR1) gene, part of a haplotype previously linked to altered function of the MDR1 gene product P-glycoprotein (P-gp), nonetheless results in P-gp with altered drug and inhibitor interactions. Similar mRNA and protein levels, but altered conformations, were found for wild-type and polymorphic P-gp. We hypothesize that the presence of a rare codon, marked by the synonymous polymorphism, affects the timing of cotranslational folding and insertion of P-gp into the membrane, thereby altering the structure of substrate and inhibitor interaction sites.
Science 26 January 2007 Vol. 315. no. 5811, pp. 525 - 528
Chava Kimchi-Sarfaty, Jung Mi Oh, In-Wha Kim, Zuben E. Sauna, Anna Maria Calcagno, Suresh V. Ambudkar, Michael M. Gottesman
Abstract: Synonymous single-nucleotide polymorphisms (SNPs) do not produce altered coding sequences, and therefore they are not expected to change the function of the protein in which they occur. We report that a synonymous SNP in the Multidrug Resistance 1 (MDR1) gene, part of a haplotype previously linked to altered function of the MDR1 gene product P-glycoprotein (P-gp), nonetheless results in P-gp with altered drug and inhibitor interactions. Similar mRNA and protein levels, but altered conformations, were found for wild-type and polymorphic P-gp. We hypothesize that the presence of a rare codon, marked by the synonymous polymorphism, affects the timing of cotranslational folding and insertion of P-gp into the membrane, thereby altering the structure of substrate and inhibitor interaction sites.
Wednesday, January 24, 2007
BiDiL: is it really an African thing?
 Here's an argument against BiDil and a response from the FDA.
Here's an argument against BiDil and a response from the FDA.BiDil for heart failure in black patients: implications of the U.S. Food and Drug Administration approval.
Ann Intern Med. 2007 Jan 2;146(1):52-6. Bibbins-Domingo K, Fernandez A.
Abstract: In 2005, the combination of hydralazine hydrochloride and isosorbide dinitrate was approved by the U.S. Food and Drug Administration (FDA) for treating heart failure in black patients. In departing from its long history of approving drugs for general clinical indications without regard to demographic classification, the FDA cited the need to address racial disparities in health as an important contributor to their decision. The authors argue that this decision, although perhaps well-intentioned, was based on flawed scientific interpretation of trial results that claimed differential drug response by race and ignored the considerable literature on the cause of racial disparities in health and health care. Because of its potential impact on future drug approvals, the FDA's decision is a setback in the scientific and policy discourse on medical therapeutics and race and specifically hinders the efforts aimed at eliminating health and health care disparities.
BiDil for heart failure in black patients: The U.S. Food and Drug Administration perspective
Temple R, Stockbridge NL.
Ann Intern Med. 2007 Jan 2;146(1):57-62
Abstract: Critics of the U.S. Food and Drug Administration (FDA) approval of the fixed combination of hydralazine hydrochloride, 37.5 mg, and isosorbide dinitrate, 20 mg, for treating heart failure in black patients have suggested that data were insufficient to distinguish treatment effects in black and white people; that distinctions based on race, rather than pathophysiology, were scientifically unreasonable; and that a "race-based" approval could be a commercial ploy to avoid a more expensive and prolonged full evaluation of a drug. The criticisms acknowledge that data supporting the approval came from a well-designed clinical trial in which self-identified black patients with heart failure who took hydralazine hydrochloride-isosorbide dinitrate with standard therapy experienced a statistically significant 43% (95% CI, 11% to 63%) reduction in mortality compared with those who took only the standard therapy. The criticisms do not always recognize that the decision to conduct the trial in only black patients reflected careful analyses of 2 previous trials in racially mixed patient populations that compared hydralazine hydrochloride-isosorbide dinitrate with placebo or with enalapril. Both trials showed little or no overall effect of hydralazine hydrochloride-isosorbide dinitrate in the mostly white patient population but hinted at a substantial effect in subsets of black patients. Perhaps most critically, the criticisms do not appreciate the urgency of strong scientific evidence of a substantial survival benefit in black patients. A serious attempt to avoid race-based approval by mandating study of a mixed population to identify a possible white patient-responder subset, particularly without a plausible hypothesis as to what that subset might be, would have required years of work, many thousands of patients, and wholly unreasonable delay in approval of a treatment whose effectiveness had been well-documented in the group for which it was intended.
Tuesday, January 23, 2007
Genetic ancestry and body composition
Nothing too earth shattering here.. another study showing that in African Americans, European ancestry correlates positively with lower BMD (bone mass density) --although this correlation was much more pronounced among women, longer trunk length, shorter leg length, and increased body fat.
The conclusion in the abstract is good: "These results indicate that some population differences in body composition may be due to population-specific allele frequencies, suggesting the utility of admixture mapping for identifying susceptibility genes for osteoporosis, sarcopenia, and obesity."
Genetic markers for ancestry are correlated with body composition traits in older African Americans
Osteoporos Int. 2007 Jan 18
Shaffer JR, Kammerer CM, Reich D, McDonald G, Patterson N, Goodpaster B, Bauer DC, Li J, Newman AB, Cauley JA, Harris TB, Tylavsky F, Ferrell RE, Zmuda JM; for the Health ABC study.
Abstract: Individual-specific percent European ancestry was assessed in 1,277 African Americans. We found significant correlations between proportion of European ancestry and several musculoskeletal traits, indicating that admixture mapping may be a useful strategy for locating genes affecting these traits. INTRODUCTION: Genotype data for admixed populations can be used to detect chromosomal regions influencing disease risk if allele frequencies at disease-related loci differ between parental populations. We assessed evidence for differentially distributed alleles affecting bone and body composition traits in African Americans. METHODS: Bone mineral density (BMD) and body composition data were collected for 1,277 African and 1,790 European Americans (aged 70-79). Maximum likelihood methods were used to estimate individual-specific percent European ancestry for African Americans genotyped at 37 ancestry-informative genetic markers. Partial correlations between body composition traits and percent European ancestry were calculated while simultaneously adjusting for the effects of covariates. RESULTS: Percent European ancestry (median = 18.7%) in African Americans was correlated with femoral neck BMD in women (r = -0.18, p < 10−5) and trabecular spine BMD in both sexes (r = −0.18, p < 10−5) independently of body size, fat, lean mass, and other covariates. Significant associations of European ancestry with appendicular lean mass (r = −0.19, p < 10−10), total lean mass (r = −0.12, p < 10−4), and total body fat (r = 0.09, p < 0.002) were also observed for both sexes. Conclusions These results indicate that some population differences in body composition may be due to population-specific allele frequencies, suggesting the utility of admixture mapping for identifying susceptibility genes for osteoporosis, sarcopenia, and obesity.
The conclusion in the abstract is good: "These results indicate that some population differences in body composition may be due to population-specific allele frequencies, suggesting the utility of admixture mapping for identifying susceptibility genes for osteoporosis, sarcopenia, and obesity."
Genetic markers for ancestry are correlated with body composition traits in older African Americans
Osteoporos Int. 2007 Jan 18
Shaffer JR, Kammerer CM, Reich D, McDonald G, Patterson N, Goodpaster B, Bauer DC, Li J, Newman AB, Cauley JA, Harris TB, Tylavsky F, Ferrell RE, Zmuda JM; for the Health ABC study.
Abstract: Individual-specific percent European ancestry was assessed in 1,277 African Americans. We found significant correlations between proportion of European ancestry and several musculoskeletal traits, indicating that admixture mapping may be a useful strategy for locating genes affecting these traits. INTRODUCTION: Genotype data for admixed populations can be used to detect chromosomal regions influencing disease risk if allele frequencies at disease-related loci differ between parental populations. We assessed evidence for differentially distributed alleles affecting bone and body composition traits in African Americans. METHODS: Bone mineral density (BMD) and body composition data were collected for 1,277 African and 1,790 European Americans (aged 70-79). Maximum likelihood methods were used to estimate individual-specific percent European ancestry for African Americans genotyped at 37 ancestry-informative genetic markers. Partial correlations between body composition traits and percent European ancestry were calculated while simultaneously adjusting for the effects of covariates. RESULTS: Percent European ancestry (median = 18.7%) in African Americans was correlated with femoral neck BMD in women (r = -0.18, p < 10−5) and trabecular spine BMD in both sexes (r = −0.18, p < 10−5) independently of body size, fat, lean mass, and other covariates. Significant associations of European ancestry with appendicular lean mass (r = −0.19, p < 10−10), total lean mass (r = −0.12, p < 10−4), and total body fat (r = 0.09, p < 0.002) were also observed for both sexes. Conclusions These results indicate that some population differences in body composition may be due to population-specific allele frequencies, suggesting the utility of admixture mapping for identifying susceptibility genes for osteoporosis, sarcopenia, and obesity.
Monday, January 22, 2007
U-turns in science
I was inspired to deviate form my usual posting on a specific paper, and to list some major "u-turns" in science recently:
- Group selection - at first a big no-no, now is become more widely accepted (but called multilevel selection).
- Population differences - AAPA said no biological basis for racial differences, now a rapidly growing field, in anthropology and medicine.
- Multiregional theory. Out of Africa with complete replacement was assumed to be "more correct" for a while, now seeing more evidence of considerable admixture/continuity
Saturday, January 20, 2007
Lactase persistence / Cystic fibrosis tradeoff
 Is the high rate of Cystic Fibrosis a by-product of lactose tolerance in Northern Europeans? This new paper in the European Journal of Human Genetics (it's open access) proposes this and provides some data to support it.
Is the high rate of Cystic Fibrosis a by-product of lactose tolerance in Northern Europeans? This new paper in the European Journal of Human Genetics (it's open access) proposes this and provides some data to support it.Cystic fibrosis and lactase persistence: a possible correlation (FREE)
Guido Modiano, Bianca M Ciminelli and Pier F Pignatti
Eur J Hum Genet advance online publication, December 20, 2006
Apparently, it has long been assumed that there was a heterozygote advantage that maintained the lethal recessive CFTR allele. Here, the authors propose that this advantage consisted of "a resistance to lactose-caused diarrhea in populations that adopted such a habit while they were still lactose intolerant."
First they argue that the high CF allele frequency in Europe (.02) compared to Africa (.01) and Far East Asia (.002) is better explained by a '' 'Europe-restricted' heterozygous advantage" than by a 'Europe-restricted' high mutation rate". (Are these really the only two possible hypotheses?).
As for what theis heterozygote advantage might consist of: "the CFTR protein is the Cl- ion channel responsible for the displacement of ... water from the mucosa epithelial cells into the respiratory and intestinal lumen."
Their time course of events:
- Initially all humans were lactose intolerant and the cumulative CF gene frequency was maintained at a value of about 0.002 by a
+
CF
4
10-6 (as in Japan).
- 5000–10 000 years ago cattle-breeding and dairy milk post-weaning diet started,19, 20 resulting in a stringent selective pressure.
- The onset of an 'emergency' adaptive response consisting of an immediate expansion of the already available CF alleles (the antiquity of the F508del allele has been estimated to be 52,000 years21). They would have attained and maintained a frequency unknown, but possibly higher than the present one, resulting from a balance between the advantage of being more resistant to the adverse consequences of being unable to digest lactose (hence to benefit from an important food as the milk) and a heavy segregational load.
- The onset of the not costly lactase-persistence adaptation, which attained rapidly a very high frequency (up to fixation in some European areas).14, 15
- A progressive decrease of the CF gene frequency down to the present (still high) values, which can be considered a relic of the past 'emergency' adaptation.
Friday, January 19, 2007
Uncontacted Amazonian groups
Check out Kambiz's post at Anthropology.net about a new report on the number of uncontacted Amazonian groups. The estimate is 67, up from previous estimates of 40. Apparently they know this from: "old and new discoveries of footprints, abandoned huts, and other signs of human life..."
Wednesday, January 17, 2007
Direct mutation rate estimation
In this paper, they find that the mutation rate that they are able to directly observe is several times higher than previous indiret estimates. The authors then remark that "selection against deleterious mutations may have a key role in explaining patterns of genetic variation in the genome, and help to maintain recombination and sexual reproduction."
This makes me think of the patterns of varaition in a gene like MC1R, where in Africa there must have been strong selection against any deleterious mutations, and as soon as the selection environment changed (Eurasia) all kinds of deleterious mutations pop up and are selected for.
Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila
Nature 445, 82-85 (4 January 2007)
Cathy Haag-Liautard, Mark Dorris, Xulio Maside, Steven Macaskill, Daniel L. Halligan, Brian Charlesworth and Peter D. Keightley
Abstract: Spontaneous mutations are the source of genetic variation required for evolutionary change, and are therefore important for many aspects of evolutionary biology. For example, the divergence between taxa at neutrally evolving sites in the genome is proportional to the per nucleotide mutation rate, u (ref. 1), and this can be used to date speciation events by assuming a molecular clock. The overall rate of occurrence of deleterious mutations in the genome each generation (U) appears in theories of nucleotide divergence and polymorphism2, the evolution of sex and recombination3, and the evolutionary consequences of inbreeding2. However, estimates of U based on changes in allozymes4 or DNA sequences5 and fitness traits are discordant6, 7, 8. Here we directly estimate u in Drosophila melanogaster by scanning 20 million bases of DNA from three sets of mutation accumulation lines by using denaturing high-performance liquid chromatography9. From 37 mutation events that we detected, we obtained a mean estimate for u of 8.4 10-9 per generation. Moreover, we detected significant heterogeneity in u among the three mutation-accumulation-line genotypes. By multiplying u by an estimate of the fraction of mutations that are deleterious in natural populations of Drosophila10, we estimate that U is 1.2 per diploid genome. This high rate suggests that selection against deleterious mutations may have a key role in explaining patterns of genetic variation in the genome, and help to maintain recombination and sexual reproduction.
Fours Stone Hearth VII
Check out 4SH VII, the Anthro Blog Carnival, this time at Aardvarchaeology, which includes an entry from yours truly (see previous post).
Tuesday, January 16, 2007
Persistence hunting
 The possibility that endurance running was an important part of the human niche is quite intriguing to me, especially given the genes (ACTN-3, GDF8, PPAR, PGC1, and others) that we could examine this hypothesis with.
The possibility that endurance running was an important part of the human niche is quite intriguing to me, especially given the genes (ACTN-3, GDF8, PPAR, PGC1, and others) that we could examine this hypothesis with.Here are some excerpts from this recent paper in Current Anthropology (link and abstract are below) that describes the effectiveness of persistence hunting (running prey into exhaustion) of !Xo and /Gwi hunters in Botswana. I thought this paper was really interesting. It goes into the details of how such hunts are conducted, compares it to other methods of hunting with actual return rate data, and goes into the unique human physiology that allows such persistence hunting. There's also an evolutionary hypothesis elaboration in the appendix, and a video too!!!
On the two hypotheses for endurance running:
Two possible hypotheses are given for why early Homo would have needed to run long distances. One hypothesis is scavenging. Competition to reach carcasses before other scavengers would have increased the fitness benefits of features that improve endurance running capabilities. Another hypothesis, presented by Carrier (1984), is that early hominin hunters used endurance running to run some mammals to exhaustion.
...and what the author finds:
Data from observations of !Xo and /Gwi hunters of the central Kalahari in Botswana presented here suggest that persistence hunting was a very efficient method under certain conditions. Compared with other forms of hunting, it may have been one of the most efficient.
the basics of how they do it:
The hunt takes place during the hottest time of the day, with maximum temperatures of about 39–42°C. Before starting, the hunters drink as much water as they can. Then they run up to the animal, which quickly flees, and track its footprints at a running pace. Meanwhile, the animal will have stopped to rest in the shade. The hunters must find the animal and chase it before it has rested long enough. This process is repeated until the animal is run to exhaustion.
..and part of their conclusion:
Compared with other hunting methods, persistence hunting is, given the right conditions, an effective method with a relatively good success rate and meat yield. The data presented suggest that it produces a higher meat yield than hunting with bow and arrow, clubs and spears, or springhare probes and about the same as snaring. Only hunting with dogs produces a significantly higher meat yield.
Persistence Hunting by Modern Hunter-Gatherers
CURRENT ANTHROPOLOGY Volume 47, Number 6, December 2006
Louis Liebenberg
Abstract: Endurance running may be a derived capability of the genus Homo and may have been instrumental in the evolution of the human body form. Two hypotheses have been presented to explain why early Homo would have needed to run long distances: scavenging and persistence hunting. Persistence hunting takes place during the hottest time of the day and involves chasing an animal until it is run to exhaustion. A critical factor is the fact that humans can keep their bodies cool by sweating while running. Another critical factor is the ability to track down an animal. Endurance running may have had adaptive value not only in scavenging but also in persistence hunting. Before the domestication of dogs, persistence hunting may have been one of the most efficient forms of hunting and may therefore have been crucial in the evolution of humans.
Costs of Carnivory
The Costs of Carnivory
Chris Carbone, Amber Teacher, J. Marcus Rowcliffe
PLoS Biology January 16, 2007
Abstract: Mammalian carnivores fall into two broad dietary groups: smaller carnivores (<20>20 kg) that specialize in feeding on large vertebrates. We develop a model that predicts the mass-related energy budgets and limits of carnivore size within these groups. We show that the transition from small to large prey can be predicted by the maximization of net energy gain; larger carnivores achieve a higher net gain rate by concentrating on large prey. However, because it requires more energy to pursue and subdue large prey, this leads to a 2-fold step increase in energy expenditure, as well as increased intake. Across all species, energy expenditure and intake both follow a three-fourths scaling with body mass. However, when each dietary group is considered individually they both display a shallower scaling. This suggests that carnivores at the upper limits of each group are constrained by intake and adopt energy conserving strategies to counter this. Given predictions of expenditure and estimates of intake, we predict a maximum carnivore mass of approximately a ton, consistent with the largest extinct species. Our approach provides a framework for understanding carnivore energetics, size, and extinction dynamics.
Chris Carbone, Amber Teacher, J. Marcus Rowcliffe
PLoS Biology January 16, 2007
Abstract: Mammalian carnivores fall into two broad dietary groups: smaller carnivores (<20>20 kg) that specialize in feeding on large vertebrates. We develop a model that predicts the mass-related energy budgets and limits of carnivore size within these groups. We show that the transition from small to large prey can be predicted by the maximization of net energy gain; larger carnivores achieve a higher net gain rate by concentrating on large prey. However, because it requires more energy to pursue and subdue large prey, this leads to a 2-fold step increase in energy expenditure, as well as increased intake. Across all species, energy expenditure and intake both follow a three-fourths scaling with body mass. However, when each dietary group is considered individually they both display a shallower scaling. This suggests that carnivores at the upper limits of each group are constrained by intake and adopt energy conserving strategies to counter this. Given predictions of expenditure and estimates of intake, we predict a maximum carnivore mass of approximately a ton, consistent with the largest extinct species. Our approach provides a framework for understanding carnivore energetics, size, and extinction dynamics.
Hispanic genetic admixture and breast cancer
- They only used 15 ancestry informative markers here and find no association between admixture and 'breast cancer risk" (don't know how they define this).
- Genetic Admixture among Hispanics and Candidate Gene Polymorphisms: Potential for Confounding in a Breast Cancer Study?
Sweeney C, Wolff RK, Byers T, Baumgartner KB, Giuliano AR, Herrick JS, Murtaugh MA, Samowitz WS, Slattery ML.
Monday, January 15, 2007
Eur-Afr Gene Expression differences
Check out agnostic's post at GNXP about another paper in AJHG about gene expression differences between Europeans and Africans. They find smaller differences (~17% of genes differentially expressed) between Eur and Afr than the recent study found between Europeans and Asians (23%).
This raises all kinds of questions (although the latest study has a very small sample compared with the Eur-As one) as to what we consider to be the major split in humans. Many studies show that Africans vs. the rest is the major split, but this and other studies have found the major split to be East Asians vs. the rest. I'm thinking mainly here of the Rosenberg et al. paper in Science a few years back where they used STRUCTURE on ~344 microsatellites and find that if K=2, those two pops are East Asians and the rest.
This raises all kinds of questions (although the latest study has a very small sample compared with the Eur-As one) as to what we consider to be the major split in humans. Many studies show that Africans vs. the rest is the major split, but this and other studies have found the major split to be East Asians vs. the rest. I'm thinking mainly here of the Rosenberg et al. paper in Science a few years back where they used STRUCTURE on ~344 microsatellites and find that if K=2, those two pops are East Asians and the rest.
Thursday, January 11, 2007
Amplifying ancient DNA
For those of you planning on excavating some fossil bones and extracting DNA:
Freshly excavated fossil bones are best for amplification of ancient DNA
Mélanie Pruvost, Reinhard Schwarz, Virginia Bessa Correia, Sophie Champlot, Séverine Braguier, Nicolas Morel, Yolanda Fernandez-Jalvo, Thierry Grange, and Eva-Maria Geigl
PNAS Published online before print January 8, 2007
Abstract: Despite the enormous potential of analyses of ancient DNA for phylogeographic studies of past populations, the impact these analyses, most of which are performed with fossil samples from natural history museum collections, has been limited to some extent by the inefficient recovery of ancient genetic material. Here we show that the standard storage conditions and/or treatments of fossil bones in these collections can be detrimental to DNA survival. Using a quantitative paleogenetic analysis of 247 herbivore fossil bones up to 50,000 years old and originating from 60 different archeological and paleontological contexts, we demonstrate that freshly excavated and nontreated unwashed bones contain six times more DNA and yield twice as many authentic DNA sequences as bones treated with standard procedures. This effect was even more pronounced with bones from one Neolithic site, where only freshly excavated bones yielded results. Finally, we compared the DNA content in the fossil bones of one animal, a 3,200-year-old aurochs, excavated in two separate seasons 57 years apart. Whereas the washed museum-stored fossil bones did not permit any DNA amplification, all recently excavated bones yielded authentic aurochs sequences. We established that during the 57 years when the aurochs bones were stored in a collection, at least as much amplifiable DNA was lost as during the previous 3,200 years of burial. This result calls for a revision of the postexcavation treatment of fossil bones to better preserve the genetic heritage of past life forms
Wednesday, January 10, 2007
Modeling the genetics of homosexuality
In this paper, the authors attempt to model how homosexuality can be sustained in a population.
According to them and others, three ways:
This is very much theoretical stuff, but according to the authors, has the potential to "generate testable predictions" regarding the genetics of homosexuality
Genetic models of homosexuality: generating testable predictions
Sergey Gavrilets and William R. Rice
Proceedings of the Royal Society B: Biological Sciences Volume 273, Number 1605 / December 22, 2006 Pages: 3031 - 3038
Abstract: Homosexuality is a common occurrence in humans and other species, yet its genetic and evolutionary basis is poorly understood. Here, we formulate and study a series of simple mathematical models for the purpose of predicting empirical patterns that can be used to determine the form of selection that leads to polymorphism of genes influencing homosexuality. Specifically, we develop theory to make contrasting predictions about the genetic characteristics of genes influencing homosexuality including: (i) chromosomal location, (ii) dominance among segregating alleles and (iii) effect sizes that distinguish between the two major models for their polymorphism: the overdominance and sexual antagonism models. We conclude that the measurement of the genetic characteristics of quantitative trait loci (QTLs) found in genomic screens for genes influencing homosexuality can be highly informative in resolving the form of natural selection maintaining their polymorphism.
According to them and others, three ways:
- overdominance
- sexually antagonistic selection
- kin altruism
This is very much theoretical stuff, but according to the authors, has the potential to "generate testable predictions" regarding the genetics of homosexuality
Genetic models of homosexuality: generating testable predictions
Sergey Gavrilets and William R. Rice
Proceedings of the Royal Society B: Biological Sciences Volume 273, Number 1605 / December 22, 2006 Pages: 3031 - 3038
Abstract: Homosexuality is a common occurrence in humans and other species, yet its genetic and evolutionary basis is poorly understood. Here, we formulate and study a series of simple mathematical models for the purpose of predicting empirical patterns that can be used to determine the form of selection that leads to polymorphism of genes influencing homosexuality. Specifically, we develop theory to make contrasting predictions about the genetic characteristics of genes influencing homosexuality including: (i) chromosomal location, (ii) dominance among segregating alleles and (iii) effect sizes that distinguish between the two major models for their polymorphism: the overdominance and sexual antagonism models. We conclude that the measurement of the genetic characteristics of quantitative trait loci (QTLs) found in genomic screens for genes influencing homosexuality can be highly informative in resolving the form of natural selection maintaining their polymorphism.
Posting comments
I just realized that I could adjust the setting for allowing comments from everyone. Before, I thought only registered Blogger users could post comments... so have at it if you wish.
Hispanic Paradox
A video in the New York Times Science section describes and attempts to explain the so-called "Hispanic Paradox", which is that Hispanic immigrants have higher life expectancy than any other group in America, despite lower socio-economic status:
possible reasons given:
- self selection of those willing to immigrate to the US. (see my post on hypomanic americans)
- cultural ways that promote stress free life and good habits
possible reasons given:
- self selection of those willing to immigrate to the US. (see my post on hypomanic americans)
- cultural ways that promote stress free life and good habits
Jablonksi at NYT
There's an interview and video with Nina Jablonski in the New York Times. Nothing earth-shattering or new here, but I thought I'd post it anyway.
She does say: "These early humans ran long distances in open areas", referring to why we have the "sweatiest (skin) of mammals" (not sure if she means we're just one of the sweatiest or THE sweatiest)
She does say: "These early humans ran long distances in open areas", referring to why we have the "sweatiest (skin) of mammals" (not sure if she means we're just one of the sweatiest or THE sweatiest)
Tuesday, January 09, 2007
Mating structure in the genes
Regarding the previous post on matrilocality, this paper examined differences in mtDNA and Y-chromosome diversity between Central Asian farmers (who are organized into nuclear families and practice endogamy, although the picture below shows exogamy) and pastoral populations (who are organized into lineages and practice exogamy). They don't find much difference in mtDNA diversity, but find less Y-chromosome diversity among the pastoral than the farming populations. They also find that the Uzbek groups who converted to a farming way of life about 20 generations ago show similar patterns of Y-chromosome diversity to the other farmers, indicating that "a molecular signature in Y-chromosome diversity is short-lived and can disappear within a few centuries after the disintegration of descent groups."
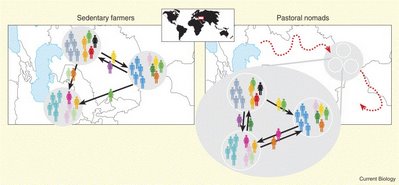 Here is the link to the commentary paper where Jobling and Balaresque assert that 70% of modern human populations practice patrilocality, a figure that I've seen disputed at least once.
Here is the link to the commentary paper where Jobling and Balaresque assert that 70% of modern human populations practice patrilocality, a figure that I've seen disputed at least once.
 Here is the link to the commentary paper where Jobling and Balaresque assert that 70% of modern human populations practice patrilocality, a figure that I've seen disputed at least once.
Here is the link to the commentary paper where Jobling and Balaresque assert that 70% of modern human populations practice patrilocality, a figure that I've seen disputed at least once.
Monday, January 08, 2007
Genetics of matrilocality, patrilocality
There is also an accompanying commentary paper by Jobling of the paper below. I haven't read either yet, but will post more on this soon.
From Social to Genetic Structures in Central Asia
Current Biology, Vol 17, 43-48, 09 January 2007
Raphaëlle Chaix, Lluís Quintana-Murci, Tatyana Hegay, Michael F. Hammer, Zahra Mobasher, Frédéric Austerlitz, and Evelyne Heyer
Abstract: Pastoral and farmer populations, who have coexisted in Central Asia since the fourth millennium B.C. [1], present not only different lifestyles and means of subsistence but also various types of social organization. Pastoral populations are organized into so-called descent groups (tribes, clans, and lineages) and practice exogamous marriages (a man chooses a bride in a different lineage or clan). In Central Asia, these descent groups are patrilineal: The children are systematically affiliated with the descent groups of the father. By contrast, farmer populations are organized into families (extended or nuclear) and often establish endogamous marriages with cousins [2, 3, 4]. This study aims at better understanding the impact of these differences in lifestyle and social organization on the shaping of genetic diversity. We show that pastoral populations exhibit a substantial loss of Y chromosome diversity in comparison to farmers but that no such a difference is observed at the mitochondrial-DNA level. Our analyses indicate that the dynamics of patrilineal descent groups, which implies different male and female sociodemographic histories, is responsible for these sexually-asymmetric genetic patterns. This molecular signature of the pastoral social organization disappears over a few centuries only after conversion to an agricultural way of life.
From Social to Genetic Structures in Central Asia
Current Biology, Vol 17, 43-48, 09 January 2007
Raphaëlle Chaix, Lluís Quintana-Murci, Tatyana Hegay, Michael F. Hammer, Zahra Mobasher, Frédéric Austerlitz, and Evelyne Heyer
Abstract: Pastoral and farmer populations, who have coexisted in Central Asia since the fourth millennium B.C. [1], present not only different lifestyles and means of subsistence but also various types of social organization. Pastoral populations are organized into so-called descent groups (tribes, clans, and lineages) and practice exogamous marriages (a man chooses a bride in a different lineage or clan). In Central Asia, these descent groups are patrilineal: The children are systematically affiliated with the descent groups of the father. By contrast, farmer populations are organized into families (extended or nuclear) and often establish endogamous marriages with cousins [2, 3, 4]. This study aims at better understanding the impact of these differences in lifestyle and social organization on the shaping of genetic diversity. We show that pastoral populations exhibit a substantial loss of Y chromosome diversity in comparison to farmers but that no such a difference is observed at the mitochondrial-DNA level. Our analyses indicate that the dynamics of patrilineal descent groups, which implies different male and female sociodemographic histories, is responsible for these sexually-asymmetric genetic patterns. This molecular signature of the pastoral social organization disappears over a few centuries only after conversion to an agricultural way of life.
Sunday, January 07, 2007
Differences in gene expression between populations
Check out p-ter's post at Gene Expression on a new paper out in Nature Genetics that compares gene expression levels between Asians and Europeans from the HapMap cell lines. As p-ter mentions, it is puzzling why they didn't include the African sample. The paper contains a list of the genes for which expression differed the most -- none were very notable to me.
Friday, January 05, 2007
Asthma: Population differences?
Barnes KC, Grant AV, Hansel NN, Gao P, Dunston GM.
Proc Am Thorac Soc. 2007;4(1):58-68
Abstract: It has been well established that genetic factors strongly affect susceptibility to asthma and its associated traits. It is less clear to what extent genetic variation contributes to the ethnic disparities observed for asthma morbidity and mortality. Individuals of African descent with asthma have more severe asthma, higher IgE levels, a higher degree of steroid dependency, and more severe clinical symptoms than individuals of European descent with asthma but relatively few studies have focused on this particularly vulnerable ethnic group. Similar underrepresentation exists for other minorities, including Hispanics. In this review, a summary of linkage and association studies in populations of African descent is presented, and the role of linkage disequilibrium in the dissection of a complex trait such as asthma is discussed. Consideration for the impact of population stratification in recently admixed populations (i.e., European, African) is essential in genetic association studies focusing on African ancestry groups. With the most recent update on the International HapMap Project, efficient selection of haplotype tagging single nucleotide polymorphisms (htSNPs) for African Americans has accelerated and efficiency of htSNPs chosen from one population to represent other continental groups (e.g., African) has been demonstrated. Cutting-edge approaches, such as genomewide association studies, admixture mapping, and phylogenetic analyses, offer new opportunities for dissecting the genetic basis for asthma in populations of African descent.
Proc Am Thorac Soc. 2007;4(1):58-68
Abstract: It has been well established that genetic factors strongly affect susceptibility to asthma and its associated traits. It is less clear to what extent genetic variation contributes to the ethnic disparities observed for asthma morbidity and mortality. Individuals of African descent with asthma have more severe asthma, higher IgE levels, a higher degree of steroid dependency, and more severe clinical symptoms than individuals of European descent with asthma but relatively few studies have focused on this particularly vulnerable ethnic group. Similar underrepresentation exists for other minorities, including Hispanics. In this review, a summary of linkage and association studies in populations of African descent is presented, and the role of linkage disequilibrium in the dissection of a complex trait such as asthma is discussed. Consideration for the impact of population stratification in recently admixed populations (i.e., European, African) is essential in genetic association studies focusing on African ancestry groups. With the most recent update on the International HapMap Project, efficient selection of haplotype tagging single nucleotide polymorphisms (htSNPs) for African Americans has accelerated and efficiency of htSNPs chosen from one population to represent other continental groups (e.g., African) has been demonstrated. Cutting-edge approaches, such as genomewide association studies, admixture mapping, and phylogenetic analyses, offer new opportunities for dissecting the genetic basis for asthma in populations of African descent.
Thursday, January 04, 2007
Marathon mice
A new paper is out where transgenic mice that over-express PGC1alpha exhibit greater endurance capacity. Science Blog link
It would be interesting to examine variation of this gene between humans and other primates and between human populations.
endurance running - important human adaptation ??
other genes related to running (with links):
in mice - PPAR
in humans - ACTN3
It would be interesting to examine variation of this gene between humans and other primates and between human populations.
endurance running - important human adaptation ??
other genes related to running (with links):
in mice - PPAR
in humans - ACTN3
Wednesday, January 03, 2007
East African mtDNA
their main selling points: whole mtDNA genome sequencing, and more East African population samples...their conclusion: they confirm the role of East African populations (especially around Tanzania?) in the origin and spread of humans to the rest of Africa and the world.
Whole mtDNA Genome Sequence Analysis of Ancient African Lineages
Mary Katherine Gonder, Holly M. Mortensen, Floyd A. Reed, Alexandra de Sousa and Sarah A. Tishkoff
Molecular Biology and Evolution Accepted for publication December 12, 2006. Abstract: Studies of human mtDNA genomes demonstrate that the root of the human phylogenetic tree occurs in Africa. Although two mtDNA lineages with an African origin (haplogroups M and N) were the progenitors of all non-African haplogroups, macrohaplogroup L (including haplogroups L0-L6) is limited to sub-Saharan Africa. Several L haplogroup lineages occur most frequently in eastern Africa (e.g., L0a, L0f, L5, L3g), but some are specific to certain ethnic groups, such as haplogroup lineages L0d and L0k that previously have been found nearly exclusively among southern African "click" speakers. Few studies have included multiple mtDNA genome samples belonging to haplogroups that occur in eastern and southern Africa but are rare or absent elsewhere. This lack of sampling in eastern Africa makes it difficult to infer relationships among mtDNA haplogroups or to examine events that occurred early in human history. We sequenced 62 complete mtDNA genomes of ethnically diverse Tanzanians, southern African Khoisan speakers and Bakola Pygmies and compared them to a global pool of 226 mtDNA genomes. From these, we infer phylogenetic relationships amongst mtDNA haplogroups and estimate the time to most recent common ancestor (TMRCA) for haplogroup lineages. These data suggest that Tanzanians have high genetic diversity and possess ancient mtDNA haplogroups, some of which are either rare (L0d and L5) or absent (L0f) in other regions of Africa. We propose that a large and diverse human population(s) has persisted in eastern Africa and that eastern Africa may have been an ancient source of dispersion of modern humans both within and outside of Africa.
Whole mtDNA Genome Sequence Analysis of Ancient African Lineages
Mary Katherine Gonder, Holly M. Mortensen, Floyd A. Reed, Alexandra de Sousa and Sarah A. Tishkoff
Molecular Biology and Evolution Accepted for publication December 12, 2006. Abstract: Studies of human mtDNA genomes demonstrate that the root of the human phylogenetic tree occurs in Africa. Although two mtDNA lineages with an African origin (haplogroups M and N) were the progenitors of all non-African haplogroups, macrohaplogroup L (including haplogroups L0-L6) is limited to sub-Saharan Africa. Several L haplogroup lineages occur most frequently in eastern Africa (e.g., L0a, L0f, L5, L3g), but some are specific to certain ethnic groups, such as haplogroup lineages L0d and L0k that previously have been found nearly exclusively among southern African "click" speakers. Few studies have included multiple mtDNA genome samples belonging to haplogroups that occur in eastern and southern Africa but are rare or absent elsewhere. This lack of sampling in eastern Africa makes it difficult to infer relationships among mtDNA haplogroups or to examine events that occurred early in human history. We sequenced 62 complete mtDNA genomes of ethnically diverse Tanzanians, southern African Khoisan speakers and Bakola Pygmies and compared them to a global pool of 226 mtDNA genomes. From these, we infer phylogenetic relationships amongst mtDNA haplogroups and estimate the time to most recent common ancestor (TMRCA) for haplogroup lineages. These data suggest that Tanzanians have high genetic diversity and possess ancient mtDNA haplogroups, some of which are either rare (L0d and L5) or absent (L0f) in other regions of Africa. We propose that a large and diverse human population(s) has persisted in eastern Africa and that eastern Africa may have been an ancient source of dispersion of modern humans both within and outside of Africa.
Tuesday, January 02, 2007
A consensus on the evolution of hominid diets?
 What I've been looking for!! (kind of). This review by John Shea of a conference (in the latest issue of Evolutionary Anthropology) held this past May at The Max Planck Institute for Evolutionary Anthropology. It was entitled "The Evolution of Hominid Diets: Integrating Approaches to the Study of Paleolithic Subsistence".
What I've been looking for!! (kind of). This review by John Shea of a conference (in the latest issue of Evolutionary Anthropology) held this past May at The Max Planck Institute for Evolutionary Anthropology. It was entitled "The Evolution of Hominid Diets: Integrating Approaches to the Study of Paleolithic Subsistence".Some of the more interesting tidbits:
"Inception of Meat-Eating":
Evidence from the earliest Homo fossils from 2.5 Mya, indicating a mixed woodland/savanna habitat, suggesting many "opportunities for hunting and scavenging" along with evidence of reduction in molar size, and encephalization "underwritten by higher qualtiy diets"... also some biogeochemical evidence of increased meat eating in Homo.
Scavenging versus Hunting:
Not much here, except a consensus that there was "complex variation in subsistence strategies"
 Resource Intensification:
Resource Intensification:"The Upper Paleolithic subsistence strategy dealt with high risks and costs by reduced residential mobility, increased investment in technology, and broader prey spectra, particularly the in-bulk collection of what would otherwise be low-ranked resources (for example, birds, fish and shellfish)."
Mismatches among lines of evidence: projectiles and fishing:
projectiles: who knows?
fish: "fish are a major source of long-chain fatty acids essential to brain growth", and fishing evidence appears in Upper Paleolithic Homo sapiens, but not Neanderthals.
check out this Max Planck website for more information, including names of participants and abstracts.
Using PCA instead of STRUCTURE
This paper is pretty technical and most of it flew over my head but this paragraph puts it into perspective:
"We can only uncover structure in the samples being analyzed. As pointed out in [39], the sampling strategy can affect the apparent structure. Rosenberg et al. [29] give a detailed discussion of the issue, and of the question of whether clines or clusters are a better description of human genetic variation. However, our “axes of variation” are likely to be relatively robust to this cline/cluster controversy. If there is a genetic cline running across a continent, and we sample two populations at the extremes, then it will appear to the analyst that the two populations form two discrete clusters. However, if the sampling strategy had been more geographically uniform, the cline would be apparent. Nevertheless, the eigenvector reflecting the cline could be expected to be very similar in both cases."
Population Structure and Eigenanalysis
Nick Patterson, Alkes L. Price, David Reich
PLoS Genetics December 2006, 2(12)
Abstract: Current methods for inferring population structure from genetic data do not provide formal significance tests for population differentiation. We discuss an approach to studying population structure (principal components analysis) that was first applied to genetic data by Cavalli-Sforza and colleagues. We place the method on a solid statistical footing, using results from modern statistics to develop formal significance tests. We also uncover a general “phase change” phenomenon about the ability to detect structure in genetic data, which emerges from the statistical theory we use, and has an important implication for the ability to discover structure in genetic data: for a fixed but large dataset size, divergence between two populations (as measured, for example, by a statistic like FST) below a threshold is essentially undetectable, but a little above threshold, detection will be easy. This means that we can predict the dataset size needed to detect structure.
"We can only uncover structure in the samples being analyzed. As pointed out in [39], the sampling strategy can affect the apparent structure. Rosenberg et al. [29] give a detailed discussion of the issue, and of the question of whether clines or clusters are a better description of human genetic variation. However, our “axes of variation” are likely to be relatively robust to this cline/cluster controversy. If there is a genetic cline running across a continent, and we sample two populations at the extremes, then it will appear to the analyst that the two populations form two discrete clusters. However, if the sampling strategy had been more geographically uniform, the cline would be apparent. Nevertheless, the eigenvector reflecting the cline could be expected to be very similar in both cases."
Population Structure and Eigenanalysis
Nick Patterson, Alkes L. Price, David Reich
PLoS Genetics December 2006, 2(12)
Abstract: Current methods for inferring population structure from genetic data do not provide formal significance tests for population differentiation. We discuss an approach to studying population structure (principal components analysis) that was first applied to genetic data by Cavalli-Sforza and colleagues. We place the method on a solid statistical footing, using results from modern statistics to develop formal significance tests. We also uncover a general “phase change” phenomenon about the ability to detect structure in genetic data, which emerges from the statistical theory we use, and has an important implication for the ability to discover structure in genetic data: for a fixed but large dataset size, divergence between two populations (as measured, for example, by a statistic like FST) below a threshold is essentially undetectable, but a little above threshold, detection will be easy. This means that we can predict the dataset size needed to detect structure.
Population differences in allograft failure
- Disparate Distribution of 16 Candidate Single Nucleotide Polymorphisms Among Racial and Ethnic Groups of Pediatric Heart Transplant Patients.
Girnita DM, Webber SA, Ferrell R, Burckart GJ, Brooks MM, McDade KK, Chinnock R, Canter C, Addonizio L, Bernstein D, Kirklin JK, Girnita AL, Zeevi A.
BACKGROUND.: Allograft failure in African-Americans remains higher than in Caucasians. Single nucleotide polymorphisms (SNPs) have been associated with altered allograft outcomes. METHODS.: In this multi-center study we compared SNP frequencies in 364 pediatric heart recipients from three ethnic/racial groups: Caucasian (n=243), African-American (n=39), and Hispanic (n=82). The target genes were: tumor necrosis factor-alpha, interleukin (IL)-10, IL-6, interferon (IFN)-gamma, vascular endothelial growth factor (VEGF), transforming growth factor-beta1, Fas, FasL, granzyme B, ABCB1, CYP3A5. RESULTS.: Compared to Caucasians, African-Americans exhibited a higher prevalence of genotypes associated with low expression of IFN-gamma (24% vs. 45.7%, P<0.001) and IL-10 (33% vs. 57.1%, P=0.052). African-Americans also exhibited an increased prevalence of high IL-6 (82.9% vs. 38.1%; P<0.001). VEGF -2578 C/C and -460 C/C genotypes were found more frequently in African-Americans and Hispanics as compared to Caucasians (P<0.001). G/G genotype of Fas and T/T genotype of FasL were expressed more often by African-American recipients. The prevalence of Granzyme B (-295A/G) genotype was differentially distributed in the three groups. Compared with Caucasians, African-Americans were twice as likely to carry the ABCB1 2677 G/G genotype (78.6% vs. 33.7%, P<0.0025), and they were more frequent carriers of the CYP3A5 *1/*1 genotype (35.7% vs. 0.6% in Caucasians and 7.2% in Hispanics; P<0.001). CONCLUSION.: African-Americans have a genetic background that may predispose to proinflammatory/lower regulatory environment, reduced drug exposure and immunosuppressive efficacy. In this ongoing multicenter study, these gene polymorphisms differences among ethnic/racial groups are being documented so that therapeutic strategies can be devised to optimize outcomes for pediatric transplant recipients.
Monday, January 01, 2007
Genetics of Obesity
Unraveling the Genetics of Human Obesity
David M. Mutch, Karine Clément
PLoS Genetics December 2006, v. 2(12)
Abstract: The use of modern molecular biology tools in deciphering the perturbed biochemistry and physiology underlying the obese state has proven invaluable. Identifying the hypothalamic leptin/melanocortin pathway as critical in many cases of monogenic obesity has permitted targeted, hypothesis-driven experiments to be performed, and has implicated new candidates as causative for previously uncharacterized clinical cases of obesity. Meanwhile, the effects of mutations in the melanocortin-4 receptor gene, for which the obese phenotype varies in the degree of severity among individuals, are now thought to be influenced by one's environmental surroundings. Molecular approaches have revealed that syndromes (Prader-Willi and Bardet-Biedl) previously assumed to be controlled by a single gene are, conversely, regulated by multiple elements. Finally, the application of comprehensive profiling technologies coupled with creative statistical analyses has revealed that interactions between genetic and environmental factors are responsible for the common obesity currently challenging many Westernized societies. As such, an improved understanding of the different “types” of obesity not only permits the development of potential therapies, but also proposes novel and often unexpected directions in deciphering the dysfunctional state of obesity.
Subscribe to:
Comments (Atom)
