Saturday, May 27, 2006
The genetics of the human brain
The Jewels of Our Genome: The Search for the Genomic Changes Underlying the Evolutionarily Unique Capacities of the Human Brain
James M. Sikela
PLoS Genetics, 2006, v. 2:646-655
Abstract: The recent publication of the initial sequence and analysis of the chimp genome allows us, for the first time, to compare our genome with that of our closest living evolutionary relative. With more primate genome sequences being pursued, and with other genome-wide, cross-species comparative techniques emerging, we are entering an era in which we will be able to carry out genomic comparisons of unprecedented scope and detail. These studies should yield a bounty of new insights about the genes and genomic features that are unique to our species as well as those that are unique to other primate lineages, and may begin to causally link some of these to lineage-specific phenotypic characteristics. The most intriguing potential of these new approaches will be in the area of evolutionary neurogenomics and in the possibility that the key human lineage–specific (HLS) genomic changes that underlie the evolution of the human brain will be identified. Such new knowledge should provide fresh insights into neuronal development and higher cognitive function and dysfunction, and may possibly uncover biological mechanisms for information storage, analysis, and retrieval never previously seen.
James M. Sikela
PLoS Genetics, 2006, v. 2:646-655
Abstract: The recent publication of the initial sequence and analysis of the chimp genome allows us, for the first time, to compare our genome with that of our closest living evolutionary relative. With more primate genome sequences being pursued, and with other genome-wide, cross-species comparative techniques emerging, we are entering an era in which we will be able to carry out genomic comparisons of unprecedented scope and detail. These studies should yield a bounty of new insights about the genes and genomic features that are unique to our species as well as those that are unique to other primate lineages, and may begin to causally link some of these to lineage-specific phenotypic characteristics. The most intriguing potential of these new approaches will be in the area of evolutionary neurogenomics and in the possibility that the key human lineage–specific (HLS) genomic changes that underlie the evolution of the human brain will be identified. Such new knowledge should provide fresh insights into neuronal development and higher cognitive function and dysfunction, and may possibly uncover biological mechanisms for information storage, analysis, and retrieval never previously seen.
Wednesday, May 24, 2006
IHEP (International Human Epigenome Project)
a news feature in Nature, about this project looking at the "epigenetic code":
Methylation and other alterations to DNA can significantly alter gene activity, causing inter-individual variation and sometimes disease, notably cancers. Such changes are 'epigenetic', and the term 'epigenome' refers to all the heritable biological factors other than DNA sequence that influence gene expression. Proposals for a large-scale Human Epigenome Project, modelled on the Human Genome Project, have provoked heated debate. Can this multimillion-dollar project be justified?
News Feature: Epigenetics: Unfinished symphony
To correctly 'play' the DNA score in our genome, cells must read another notation that overlays it — the epigenetic code. A global effort to decode it is now in the making, reports Jane Qiu.
-- more on this when I finish reading it.Tuesday, May 23, 2006
Population subdivision in Indian subcastes
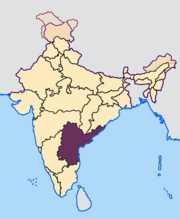
Genetic diversity within a caste population of India as measured by Y-chromosome haplogroups and haplotypes: Subcastes of the Golla of Andhra Pradesh
R. J. Mitchell, B. M. Reddy, D. Campo, T. Infantino, M. Kaps, M.H. Crawford
AJPA: July, 2006, 130:385-393
The extent of population subdivision based on 15 Y-chromosome polymorphisms was studied in seven subcastes of the Golla (Karnam, Pokanati, Erra, Doddi, Punugu, Puja, and Kurava), who inhabit the Chittoor district of southern Andhra Pradesh, India. These Golla subcastes are traditionally pastoralists, culturally homogeneous and endogamous. DNA samples from 146 Golla males were scored for seven unique event polymorphisms (UEPs) and eight microsatellites, permitting allocation of each into haplogroups and haplotypes, respectively. Genetic diversity (D) was high (range, 0.9048-0.9921), and most of the genetic variance (>91%) was explained by intrapopulation differences. Median-joining network analysis of microsatellite haplotypes demonstrated an absence of any structure according to subcaste affiliation. Superimposition of UEPs on this phylogeny, however, did create some distinct clusters, indicating congruence between haplotype and haplogroup phylogenies. Our results suggest many male ancestors for the Golla as well as for each of the subcastes. Genetic distances among the seven subcastes, based on autosomal markers (short tandem repeats and human leukocyte antigens) as well as those on the chromosome Y, indicate that the Kurava may not be a true subcaste of the Golla. Although this finding is based on a very small Kurava sample, it is in accordance with ethnohistorical accounts related by community elders. The Punugu was the first to hive off the main Golla group, and the most recently separated subcastes (Karnam, Erra, Doddi, and Pokanati) fissioned from the Puja. This phylogeny receives support from the analysis of autosomal microsatellites as well as HLA loci in the same samples. In particular, there is a significant correlation (r = 0.8569; P = 0.0097) between Y-chromosome- and autosomal STR-based distances.
some notes:
- concordance between population subdivision based on Y-chromosome analysis and population subdivision based on perceptions of elders is mixed.
- some controversy as to when the Indian caste system originated
- this population practices consanguineous marriage and village endogamy (so, no male or female biased dispersion, I guess?)
-TMRCA of these Y-chromosomes is 34, 370, lending some support to the hypothesis that the caste system originated earlier than some think.
Sunday, May 21, 2006
Do men hunt to provision or to show off?
A new paper in Current Anthropology:
Prestige or Provisioning? A Test of Foraging Goals among the Hadza
Brian Wood
Current Anthropology, 47:383-387
Tests of hypotheses concerning the foraging goals of Hadza men and women using an interview involving a hypothetical instance of foraging group formation show that most Hadza men and all Hadza women prefer to join foraging groups that ensure the greatest household provisioning advantages. Men with dependent offspring are no more likely to choose a strategy beneficial for household provisioning than men without dependent offspring. These results suggest that most Hadza men agree with women's camp preferences and value family provisioning more than broadcasting signals of their hunting ability when deciding with whom to live.
Some have argued that the predominant motivation for male hunting (especially of large game) is to aquire benefits of prestige. This paper fails to support this hypothesis as it shows that Hadza men prefer to join a camp with a lot of good hunters rather than join a group of poor hunters (where they might have higher prestige, themselves). This is similar to the question of whether to join a team of very good players (not so much individual prestige) or join a team of mediocre players (more individual prestige). I think than in this and the hunting case, mens' responses will differ based on their own hunting ability and will also be partly based on the fact that a really good hunter/player can't be good when he isn't surrounded by other equally good players/hunters. I am not sure to what degree teamwork is an important part of Hadza hunting.
final thoughts:
- two hypotheses examined in this paper are not mutually exclusive.
- prestige seems to be more of a byproduct than a primary motivator for male hunting, and is probably dependent on marital status/age/number of dependent offspring (as shown in the Ache, but not here).
Prestige or Provisioning? A Test of Foraging Goals among the Hadza
Brian Wood
Current Anthropology, 47:383-387
Tests of hypotheses concerning the foraging goals of Hadza men and women using an interview involving a hypothetical instance of foraging group formation show that most Hadza men and all Hadza women prefer to join foraging groups that ensure the greatest household provisioning advantages. Men with dependent offspring are no more likely to choose a strategy beneficial for household provisioning than men without dependent offspring. These results suggest that most Hadza men agree with women's camp preferences and value family provisioning more than broadcasting signals of their hunting ability when deciding with whom to live.
Some have argued that the predominant motivation for male hunting (especially of large game) is to aquire benefits of prestige. This paper fails to support this hypothesis as it shows that Hadza men prefer to join a camp with a lot of good hunters rather than join a group of poor hunters (where they might have higher prestige, themselves). This is similar to the question of whether to join a team of very good players (not so much individual prestige) or join a team of mediocre players (more individual prestige). I think than in this and the hunting case, mens' responses will differ based on their own hunting ability and will also be partly based on the fact that a really good hunter/player can't be good when he isn't surrounded by other equally good players/hunters. I am not sure to what degree teamwork is an important part of Hadza hunting.
final thoughts:
- two hypotheses examined in this paper are not mutually exclusive.
- prestige seems to be more of a byproduct than a primary motivator for male hunting, and is probably dependent on marital status/age/number of dependent offspring (as shown in the Ache, but not here).
Friday, May 19, 2006
Need for an evolutionary perspective in medicine
This is a short editorial from a few months back in Science by:
R. Nesse, S. Stearns & G. Omenn
Medicine Needs Evolution
Science, v. 311:1071
Areas of particular interest:
-anatomical anomalies
-infertility
-infection
-metabolic syndromes
-persistence of genes that cause certain diseases
-when is it safe to block cough, fever, diarrhea etc...?
The authors call for an incorporation of evolutionary theory in medical licensing exams, "ensure evolutionary expertise in agencies that fund biomedical research", and incorporate evol. theory into all curriculums.
...and this more recent very short letter by Joseph McInerney (Science 312:998) discusses the insights gained from "evolution theory's recognition of individual variation within populations of organisms."
He also briefly mentions the "recently announced Genes and Environment Initiative at NIH, which will investigate the interaction of genetic and environmental variations in common diseases."
R. Nesse, S. Stearns & G. Omenn
Medicine Needs Evolution
Science, v. 311:1071
Areas of particular interest:
-anatomical anomalies
-infertility
-infection
-metabolic syndromes
-persistence of genes that cause certain diseases
-when is it safe to block cough, fever, diarrhea etc...?
The authors call for an incorporation of evolutionary theory in medical licensing exams, "ensure evolutionary expertise in agencies that fund biomedical research", and incorporate evol. theory into all curriculums.
...and this more recent very short letter by Joseph McInerney (Science 312:998) discusses the insights gained from "evolution theory's recognition of individual variation within populations of organisms."
He also briefly mentions the "recently announced Genes and Environment Initiative at NIH, which will investigate the interaction of genetic and environmental variations in common diseases."
Thursday, May 18, 2006
Consumer Ethology
An interesting piece from PCMag (via TAMU Anthropology in the News) on how major corporations spend billions (yes, billions) on ethnographic research. This can involve looking at differences within and between cultures, between sexes, and across different age groups.
Wednesday, May 17, 2006
Nuclear DNA recovered from Neanderthal
John Hawks has a post on a report that Paabo et al. have been able to sequence parts of the Y-chromosome in Neanderthal remains found in a cave in Croatia. I wonder how much of the genome they will be able to amplify. What are the major questions that they might attempt to answer?
-admixture with moderns
-time since divergence (looks like 315,000 years ago, according to their analysis)
-Neanderthal genetic diversity (from the 10 or so individuals that they hope to eventually look at)
-a multitute of polymorphisms that are of interest in humans - what do they look like in Neanderthals? etc...
-admixture with moderns
-time since divergence (looks like 315,000 years ago, according to their analysis)
-Neanderthal genetic diversity (from the 10 or so individuals that they hope to eventually look at)
-a multitute of polymorphisms that are of interest in humans - what do they look like in Neanderthals? etc...
Tuesday, May 16, 2006
More on mutations in humans
From a paper in Nature in 1999 (397:344-347), by the same authors as the paper below on fitness effects of mutations:
High genomic deleterious mutation rates in hominids
Adam Eyre-Walker, Peter D. Keightley
It has been suggested that humans may suffer a high genomic deleterious mutation rate,. Here we test this hypothesis by applying a variant of a molecular approach to estimate the deleterious mutation rate in hominids from the level of selective constraint in DNA sequences. Under conservative assumptions, we estimate that an average of 4.2 amino-acid-altering mutations per diploid per generation have occurred in the human lineage since humans separated from chimpanzees. Of these mutations, we estimate that at least 38% have been eliminated by natural selection, indicating that there have been more than 1.6 new deleterious mutations per diploid genome per generation. Thus, the deleterious mutation rate specific to protein-coding sequences alone is close to the upper limit tolerable by a species such as humans that has a low reproductive rate, indicating that the effects of deleterious mutations may have combined synergistically. Furthermore, the level of selective constraint in hominid protein-coding sequences is atypically low. A large number of slightly deleterious mutations may therefore have become fixed in hominid lineages.
High genomic deleterious mutation rates in hominids
Adam Eyre-Walker, Peter D. Keightley
It has been suggested that humans may suffer a high genomic deleterious mutation rate,. Here we test this hypothesis by applying a variant of a molecular approach to estimate the deleterious mutation rate in hominids from the level of selective constraint in DNA sequences. Under conservative assumptions, we estimate that an average of 4.2 amino-acid-altering mutations per diploid per generation have occurred in the human lineage since humans separated from chimpanzees. Of these mutations, we estimate that at least 38% have been eliminated by natural selection, indicating that there have been more than 1.6 new deleterious mutations per diploid genome per generation. Thus, the deleterious mutation rate specific to protein-coding sequences alone is close to the upper limit tolerable by a species such as humans that has a low reproductive rate, indicating that the effects of deleterious mutations may have combined synergistically. Furthermore, the level of selective constraint in hominid protein-coding sequences is atypically low. A large number of slightly deleterious mutations may therefore have become fixed in hominid lineages.
Fitness effects of mutations in humans
I'm not at all familiar with the methods in this paper, but the conclusion sounds interesting, namely that "it will be difficult to locate the majority of mutations involved in genetic disease unless the disease is completely un-associated with fitness, or some of the mutations have been subject to positive selection."
The Distribution of Fitness Effects of New Deleterious Amino Acid Mutations in Humans
Adam Eyre-Walker, Meg Woolfit, Ted Phelps
Genetics, March 17, 2006, Epub ahead of print.
Abstract: The distribution of fitness effects of new mutations is a fundamental parameter in genetics. Here we present a new method by which the distribution can be estimated. The method is fairly robust to changes in population size and admixture, and it can be corrected for any residual effects if a model of the demography is available . We apply the method to extensively sampled single nucleotide polymorphism data from humans and estimate the distribution of fitness effects for amino acid changing mutations. We show that a gamma distribution with a shape parameter of 0.23 provides a good fit to the data and we estimate that more than 50% of mutations are likely to have mild effects, such that they reduce fitness by between 1/1000 and 1/10. We also infer that fewer than 15% of new mutations are likely to have strongly deleterious effects. We estimate that on average a non-synonymous mutation reduces fitness by ~4.3% and that the average strength of selection acting against a non-synonymous polymorphism is ~9 x 10-5. We argue that the relaxation of natural selection due to modern medicine and reduced variance in family size is not likely to lead to a rapid decline in genetic quality, but that it will be very difficult to locate most of the genes involved in complex genetic diseases.
The Distribution of Fitness Effects of New Deleterious Amino Acid Mutations in Humans
Adam Eyre-Walker, Meg Woolfit, Ted Phelps
Genetics, March 17, 2006, Epub ahead of print.
Abstract: The distribution of fitness effects of new mutations is a fundamental parameter in genetics. Here we present a new method by which the distribution can be estimated. The method is fairly robust to changes in population size and admixture, and it can be corrected for any residual effects if a model of the demography is available . We apply the method to extensively sampled single nucleotide polymorphism data from humans and estimate the distribution of fitness effects for amino acid changing mutations. We show that a gamma distribution with a shape parameter of 0.23 provides a good fit to the data and we estimate that more than 50% of mutations are likely to have mild effects, such that they reduce fitness by between 1/1000 and 1/10. We also infer that fewer than 15% of new mutations are likely to have strongly deleterious effects. We estimate that on average a non-synonymous mutation reduces fitness by ~4.3% and that the average strength of selection acting against a non-synonymous polymorphism is ~9 x 10-5. We argue that the relaxation of natural selection due to modern medicine and reduced variance in family size is not likely to lead to a rapid decline in genetic quality, but that it will be very difficult to locate most of the genes involved in complex genetic diseases.
The advantages of publishing in open access journals
In the new issue of PLoS Biology (open access). I had trouble getting the end of the abstact for some reason so you might just want to click on the link (there is more below, after the abstract):
Citation Advantages of Open Access Articles
Gunther Eysenbach
PLoS Biology, v.4, May 2006, p.692-698
Abstract:"Open access (OA) to the research literature has the potential to accelerate recognition and dissemination of research findings, but its actual effects are controversial. This was a longitudinal bibliometric analysis of a cohort of OA and non-OA articles published between June 8, 2004, and December 20, 2004, in the same journal (PNAS: Proceedings of the National Academy of Sciences). Article characteristics were extracted, and citation data were compared between the two groups at three different points in time: at "quasi-baseline" (December 2004, 0-6 mo after publication), in April 2005 (4-10 mo after publication), and in October 2005 (10-16 mo after publication). Potentially confounding variables, including number of authors, authors' lifetime publication count and impact, submission track, country of corresponding author, funding organization, and discipline, were adjusted for in logistic and linear multiple regression models. A total of 1,492 original research articles were analyzed: 212 (14.2% of all articles) were OA articles paid by the author, and 1,280 (85.8%) were non-OA articles. In April 2005 (mean 206 d after publication), 627 (49.0%) of the non-OA articles versus 78 (36.8%) of the OA articles were not cited (relative risk = 1.3 [95% Confidence Interval: 1.1-1.6]; p = 0.001). 6 mo later (mean 288 d after publication), non-OA articles were still more likely to be uncited (non-OA: 172 [13.6%], OA: 11 [5.2%]; relative risk = 2.6 [1.4-4.7]; p<0.001).the sd =" 2.5]" sd =" 2.0];" z =" 3.123;" p =" 0.002;" sd =" 10.4]" sd =" 4.9];" z =" 4.058;" ratio =" 2.1">
I find it surprising that there would be a significant increase in citations from papers in an open-access journal, since it is very probable that people who would be citing papers would already have access through their research institution.
The authors seem to have included a substantial number of controls, and also briefly examined self-archived journals, such as those that are on an author's website or can be obtained through Google or other internet site.
The authors briefly discuss in the conclusion below how more papers/journals might become open access:
"OA journals and hybrid journals like PNAS, as well as traditional publishers like Blackwell Publishing (“Online Open”), Oxford University Press (“Oxford Open”), and Springer (“Springer Open Choice”) are now offering authors an immediate OA option if the author pays a fee. Researchers, publishers, and policymakers confronted with the question of whether or not to invest in OA publishing have reason to believe that OA accelerates scientific advancement and knowledge translation of research into practice. While more work remains to be done to evaluate citation patterns over longer periods of time and in different fields and journals, this study provides evidence and new arguments for scientists and granting agencies to invest money into article processing fees to cover the costs of OA publishing. It also provides an incentive for publishers seeking to increase their impact factor to offer an OA option.
Citation Advantages of Open Access Articles
Gunther Eysenbach
PLoS Biology, v.4, May 2006, p.692-698
Abstract:"Open access (OA) to the research literature has the potential to accelerate recognition and dissemination of research findings, but its actual effects are controversial. This was a longitudinal bibliometric analysis of a cohort of OA and non-OA articles published between June 8, 2004, and December 20, 2004, in the same journal (PNAS: Proceedings of the National Academy of Sciences). Article characteristics were extracted, and citation data were compared between the two groups at three different points in time: at "quasi-baseline" (December 2004, 0-6 mo after publication), in April 2005 (4-10 mo after publication), and in October 2005 (10-16 mo after publication). Potentially confounding variables, including number of authors, authors' lifetime publication count and impact, submission track, country of corresponding author, funding organization, and discipline, were adjusted for in logistic and linear multiple regression models. A total of 1,492 original research articles were analyzed: 212 (14.2% of all articles) were OA articles paid by the author, and 1,280 (85.8%) were non-OA articles. In April 2005 (mean 206 d after publication), 627 (49.0%) of the non-OA articles versus 78 (36.8%) of the OA articles were not cited (relative risk = 1.3 [95% Confidence Interval: 1.1-1.6]; p = 0.001). 6 mo later (mean 288 d after publication), non-OA articles were still more likely to be uncited (non-OA: 172 [13.6%], OA: 11 [5.2%]; relative risk = 2.6 [1.4-4.7]; p<0.001).the sd =" 2.5]" sd =" 2.0];" z =" 3.123;" p =" 0.002;" sd =" 10.4]" sd =" 4.9];" z =" 4.058;" ratio =" 2.1">
I find it surprising that there would be a significant increase in citations from papers in an open-access journal, since it is very probable that people who would be citing papers would already have access through their research institution.
The authors seem to have included a substantial number of controls, and also briefly examined self-archived journals, such as those that are on an author's website or can be obtained through Google or other internet site.
The authors briefly discuss in the conclusion below how more papers/journals might become open access:
"OA journals and hybrid journals like PNAS, as well as traditional publishers like Blackwell Publishing (“Online Open”), Oxford University Press (“Oxford Open”), and Springer (“Springer Open Choice”) are now offering authors an immediate OA option if the author pays a fee. Researchers, publishers, and policymakers confronted with the question of whether or not to invest in OA publishing have reason to believe that OA accelerates scientific advancement and knowledge translation of research into practice. While more work remains to be done to evaluate citation patterns over longer periods of time and in different fields and journals, this study provides evidence and new arguments for scientists and granting agencies to invest money into article processing fees to cover the costs of OA publishing. It also provides an incentive for publishers seeking to increase their impact factor to offer an OA option.